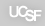
Download
FileMakerPro (.fp7):
Degrabase 1.0
(181 MB)
Excel (.xlsx):
Degrabase 1.0
(2.5 MB)

![]()
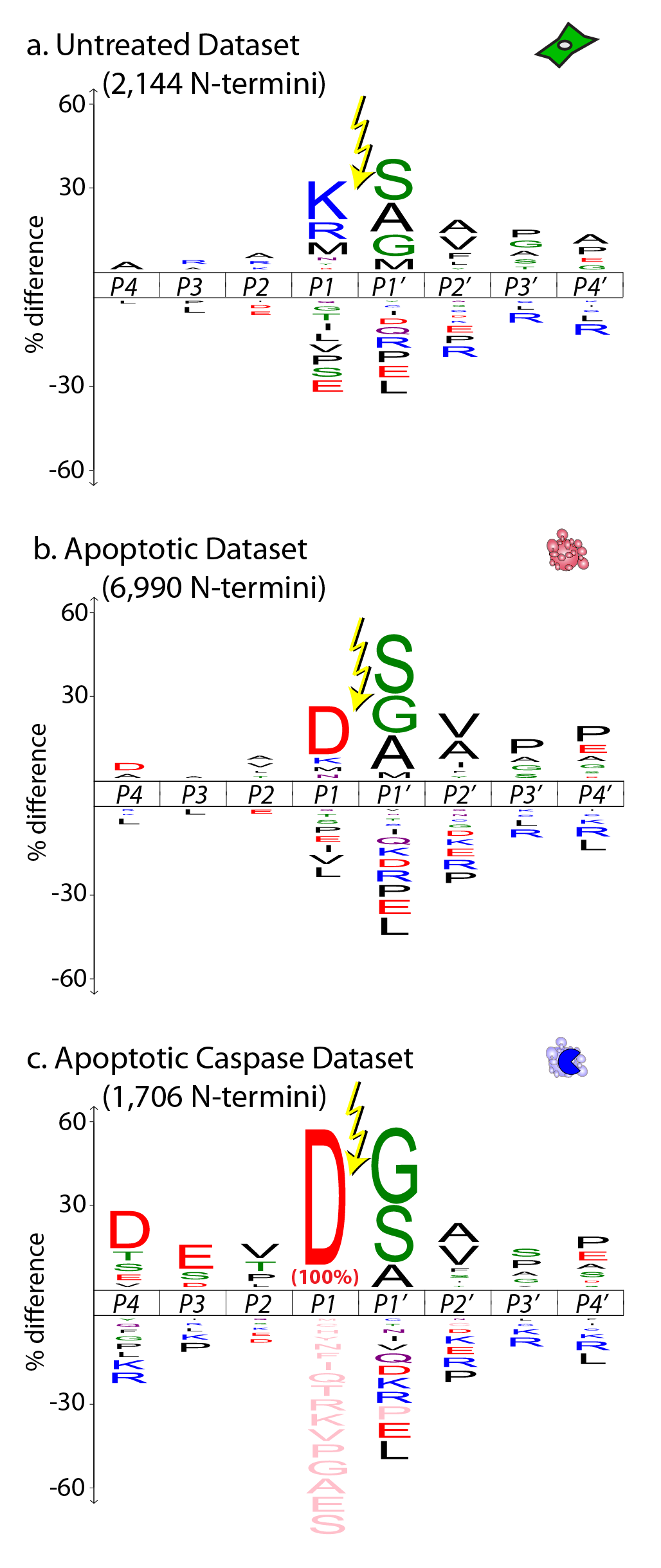
The status of the α-aminome, or all protein N-termini at a given time in a cell, can provide a wealth of information regarding protein turnover, protein trafficking, and protease signaling. The Degrabase aim to provide a non-biased description of all possible caspases substrates found in healthy and apoptotic human cells. This page was created to share with the scientific community this valuable dataset that contains over 8,000 unique α-amines detected with high confidence using mass spectrometry and our degradomics methodology.
Search
Find the official entry name of your protein of interest: http://uniprot.org
Reference
Crawford E.D.,* Seaman J.E.,* Agard N., Hsu G.W., Julien O., Mahrus S., Nguyen H., Shimbo K., Yoshihara H., Zhuang M., Chalkley R.J., Wells J.A. (2013) "The DegraBase: a database of proteolysis in healthy and apoptotic human cells." Mol. Cell. Proteomics 12. 813-824.
Wells Lab © 2011 | Site Map